What is array CGH?
A technique used in molecular biology for the detection of genomic dosage changes (deletions or duplications) at a much higher level of resolution than karyotyping.
What is a CHG array?
Array CGH (comparative genomic hybridisation) allow the detection of genomic dosage changes (deletions or duplications) with a level of resolution far superior to karyotyping.
Pros and cons of a CGH array
Advantages of a CHG array
The advantages of this technique are:
- Greater sensitivity and resolving power with respect to karyotyping to detect deletions, duplications or unbalanced rearrangements.
- Possibility of automation.
- Shorter turnaround time as no cell culture is required to obtain DNA.
Disadvantages of a CGH array
The disadvantages of this technique are that it cannot detect:
- Balanced chromosomal rearrangements, such as translocations or inversions.
- Chromosomal localisation of duplications (requires an additional FISH test).
- Low frequency mosaicism (<30% of abnormal cells).
- Some types of polyploidy such as triploidy.
- Uniparental disomy (UPD).
- Variations in repetitive regions, such as the triplet expansions of fragile X syndrome.
- Imprinting defects.
- Genetic diseases caused by point mutations or multifactorial inheritance.
Applications of a CHG array
Applications of a prenatal CHG array
Array CGH is used in prenatal diagnosis:
- For a more precise characterisation of anomalies detected by traditional methods (karyotyping).
- When structural abnormalities of the foetus are observed on ultrasound or growth retardation.
- In cases of intrauterine foetal death or stillbirth.
- At the request of the patient or physician.
Microarrays increase the detection rate of relevant chromosomal alterations in high-risk pregnancies by 5-10% and 1-2% in low-risk pregnancies. Because these techniques (as well as the exome and genome) sometimes detect more variants of uncertain significance (VUS), it is important that an expert professional interprets the data and advises the physician and/or patient.
Applications of a postnatal CGH array
It is the initial technique of choice for the study of patients with:
- Psychomotor developmental delay/intellectual disability
- Delayed growth
- Autism spectrum disorders
- Moderate to severe learning difficulties
- Dysmorphic features
- Congenital malformations of unclear aetiology
This is because they offer a higher diagnostic yield (15-20%) compared to karyotyping. The latter technique should be used as a primary indication for the study of patients with recognisable chromosomal patterns, cases with a family history of chromosomal rearrangements and in the study of infertility and/or multiple miscarriages.
What is the resolution of an array CGH?
Compared to conventional chromosome analysis, which has a detection limit of 5 to 10 Mb, array CGH can detect genomic imbalances with a diagnostic resolution of 50 to 100 kb.
CGH arrays can be designed to cover any region of interest and achieve different resolutions.
The level of resolution is determined by the size of the probe used, the genomic distance between the probes or the number of probes in the array. What does this mean?
- The smaller the DNA fragment used as a probe and the closer two consecutive probes are to each other, the higher the resolution.
- A 60k array has 60,000 probes or oligonucleotides distributed across the genome.
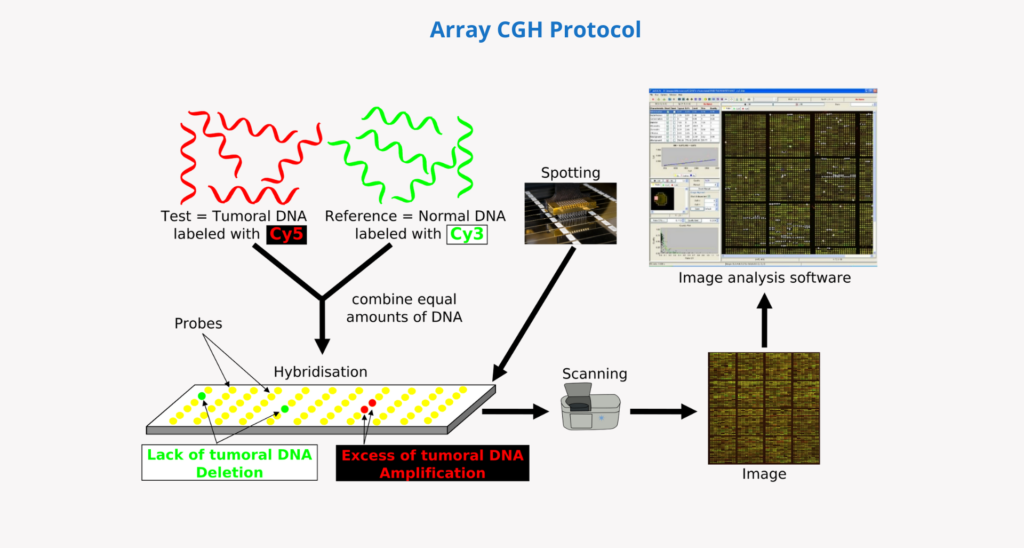
How does an array CGH work?
- First, the patient’s DNA is extracted and digested (from a sample of blood, saliva, chorionic villi, amniotic fluid or other cells or tissues).
- The same amount of patient DNA and a reference (control) DNA are then labelled with different fluorophores.
- They are then mixed and denatured from double stranded to single stranded.
- In the next step they are hybridised on a chip with immobilised DNA fragments (probes). These probes cover the human genome as uniformly as possible and can be customised according to the needs of the analysis.
- Patient and control DNA fragments compete for hybridisation with complementary sequences on the array probes.
- A high-resolution laser scanner is then used to capture and quantify the fluorescent signal intensity of each colour that has hybridised to each probe.
- Finally, the ratio of the fluorescent signal intensity in the patient’s DNA and the reference DNA is calculated for each probe in the array, obtaining information on the relative copy number of sequences in the genome in the patient compared to the reference genome. This information is interpreted by analysis software which allows the data to be assigned to the respective chromosomal region and represented as a “molecular karyotype“.