What is exome sequencing?
It is a massive sequencing technique (NGS, next generation sequencing) that allows the sequencing, in parallel, of millions of DNA fragments, covering all genes (WES, whole exome sequencing)
Massive sequencing: exome analysis
Whole exome sequencing (WES) addresses (almost) all genes in an individual, and is able to identify point mutations and other types of genetic aberrations (such as insertions and deletions) in DNA.
As with genome sequencing, its clinical utility is becoming increasingly popular. For example, recent studies report a success rate in the diagnosis of rare diseases of close to 25%.
Advantages and disadvantages of whole exome analysis
Advantages of whole exome sequencing:
The advantages of this technique are:
- It provides a high-resolution, base-by-base view of the exome.
- It can detect single nucleotide variants and insertions/deletions.
- Faster data collection than genome analysis. Generally only “only” about 25,000 variants to analyse. That is, less data to be filtered, investigated and stored.
- Currently, most of the known point mutations associated with diseases are in the coding region (exome) of the DNA.
Disadvantages of whole exome sequencing:
- Detection of CNVs (copy number variants) not as reliable as exome sequencing.
- Difficult to detect large structural variants.
- Does not detect variants in non-coding regions.
Clinical applications of whole exome analysis
Whole exome analysis covers multiple aspects of health:
- Identifying monogenic diseases (such as thalassaemia).
- Identification of the possible cause in rare diseases.
- Identifying a predisposition to develop a polygenic disease (such as type 2 diabetes mellitus).
- Identifying genetic carriers of recessive diseases, such as cystic fibrosis.
The importance of good data interpretation
Exome sequencing allows the identification of many more variants (point mutations, insertions and deletions) than other techniques, although the significance of some of this information is unknown. Since not all genetic changes affect health, it is sometimes difficult to know whether the variants identified are involved in the health of the person or disease of interest. Sometimes an identified variant is associated with a different genetic disorder that has not yet been diagnosed (called incidental or secondary findings).
This is why it is essential to have a team of experts capable of interpreting the data and answering the questions of doctors and patients.
What genetic variants can be detected by an exome?
Applications of whole exome sequencing (WES) are the detection of single nucleotide changes (SNV, single nucleotide variant or SNP, single nucleotide polymorphism) or point mutations and small copy number variations such as small insertions and deletions in coding regions (genes) of DNA.
What does coverage indicate?
The depth of coverage indicates the number of times a genome base has been sequenced. The higher the coverage, the higher the reliability of the method. In other words, the percentage of false negatives and false positives is reduced, and it is even possible to detect variants present in mosaicism.
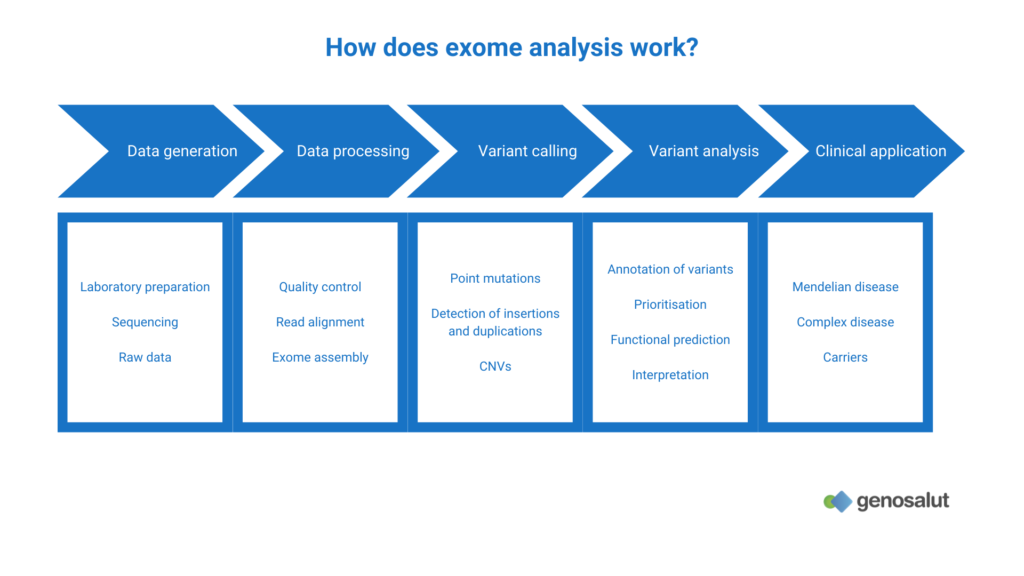
How does whole exome sequencing work?
Exome sequencing is very similar to genome sequencing. Its main difference is that it requires an enrichment step of the coding regions (genes) after preparation of the libraries and before amplification of the fragments.
That is, the exome has to be “separated” from the genome before sequencing, as it contributes only 1% of the total nuclear DNA. The basic idea of enrichment is to separate anything of interest from other substances using the difference in physicochemical properties between them.
There are two main methods to achieve exome enrichment:
- Array-based exome enrichment. It uses specific probes attached to high-density microarrays to capture the exome. Only those DNA fragments that possess a complementary sequence will be bound to the microarray.
- Enrichment of the exome in solution using biotinylated probes. Again, only those DNA fragments with a complementary sequence will be bound to the probes. The pool is enriched in desired regions by adding streptavidin microspheres that bind to the biotinylated probes. Finally, the streptavidin beads enriched with the fragments of interest are magnetically extracted.
